Eye cells to store complex data
Remember Escherichia coli? Before long, these handy bacteria might also remember you, now that Rice University synthetic biologists have won National Science Foundation (NSF) support to modify living cells to act as a memory storage device.

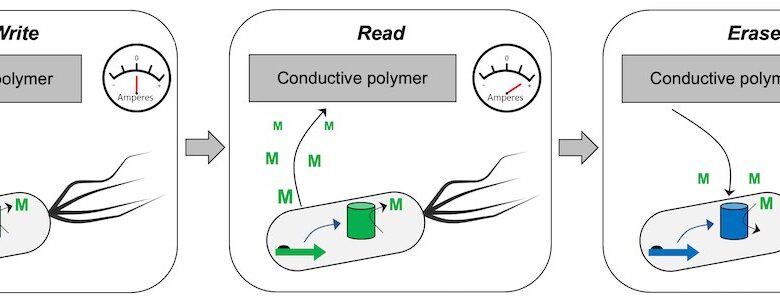
Cells turned into random access memory devices will be able to store and report data about their environment. Image credit: Silberg Lab
Agency will send $1.5 million to Rice for principal investigator to study Jonathan SilbergProfessor of Biochemistry Stewart Memorial, and his colleagues to create memories equivalent to the read-write-erase memories commonly found in computers and other devices.
These programmable cells, starting with E. coli, will be able to collect and store information about their environment for researchers to analyze later.
Silberg, director of Rice’s Systematic, Synthetic and Doctoral Biology Physics program. “While cells have the potential to be integrated with semiconductor materials to create an entirely new generation of sustainable technology that can read out information stored in biomolecules, we still There are limits to the ability to communicate between cells and materials to create useful bioelectronic devices.”
The co-investigator is Rafael Verduzcoa professor of chemical and biomolecular engineering and materials science and nanoengineering, James Chappellan assistant professor of biological sciences and bioengineering, and Caroline Ajo-Franklina professor of biological sciences and bioengineering.
“Cells can already be programmed to act as read-only memory (ROM), storing their data in DNA,” Chappell said. “Actually, biological memory can be achieved information density beyond commercial memory storage devices. However, imitation temporary access memory (RAM) and its many read-write-erase capabilities are the challenge we want to solve.”
First, instead of encoding data directly into DNA, the team encodes hundreds of bytes information in synthetic RNA biomolecules located in the cell, which will also be programmed to synthesize redox activity small molecules that carry information to and from the outside world.
Visually, the memory encoded inside these redox-active molecules can be quickly read by semiconductor polymers, equivalent to the wires and associated electronics that carry information from the memory. computer processing to the screen.
They will require an interface to read and write data. The researchers are designing printable hydrogel-based chambers containing bacteria and related electronics to read small molecules that pass through cell membranes.
This will allow cells to be programmed quickly to capture information about a variety of targets. And all this must be done without harming the cells themselves.
The RAM generated in these cells is expected to be stable for several days to weeks or even longer, although that aspect of the proposed bioelectronic devices would require tissue analysis. characterization to understand the technology’s full potential, says Silberg.
He notes that all this work will require expertise in many fields to create electrical interfaces between bacteria and materials, including RNA biology, microbial electrochemistry, biomolecular engineering. , conversion engineering and semiconductor polymers.
“At Rice, we have diverse researchers in materials science and synthetic biology who are interested in connecting advanced materials to microbial systems,” says Silberg.
Source: Rice University